New article: „Resolving complex cartilage structures in developmental biology via deep learning-based automatic segmentation of X-ray computed microtomography images“
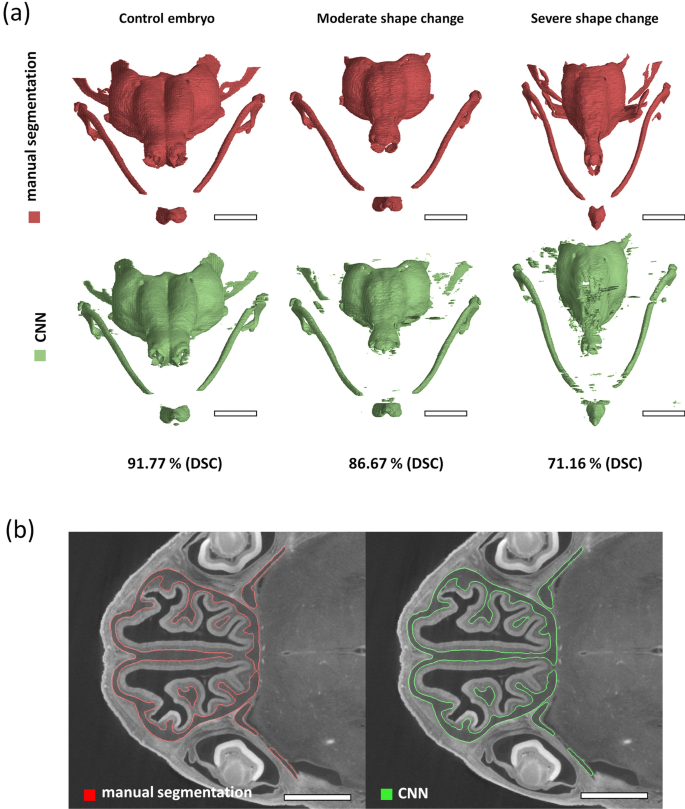
Analysis of X-ray computed tomography images in developmental biology still heavily relies on manual user input. The 3-D morphology is usually analysed by outlining the biological structures of interest by hand. This manual input can cost the researchers hundreds of hours in intensive labour, which could be better spent elsewhere.
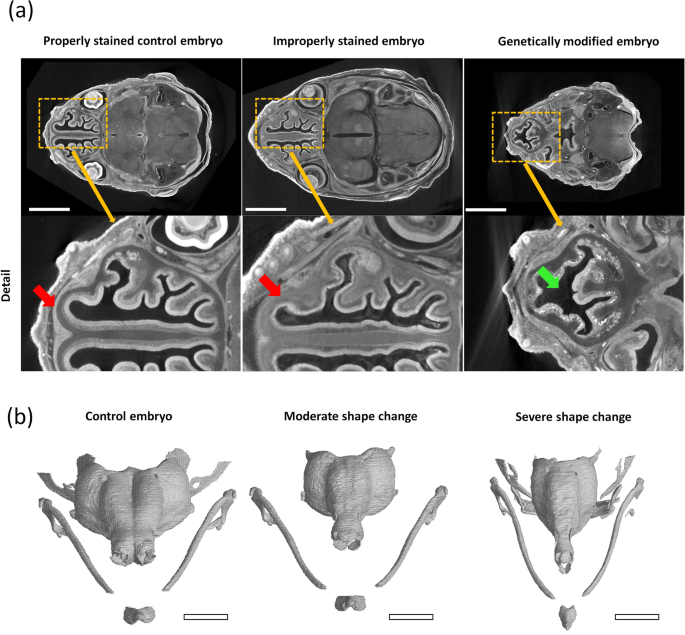
Our recently published paper investigated the possibility of offsetting this immense time investment to AI-based image processing methods. We developed a deep learning model that can produce 3-D models of the developing cartilaginous mouse embryo skull with precision nearing the accuracy of an experienced human performing the segmentation manually. More importantly, the proposed model can do this in only a fraction of the previously required time. For more information on this research on the intersection of X-ray computed tomography, developmental biology, and deep learning, see the complete publication in Scientific Reports